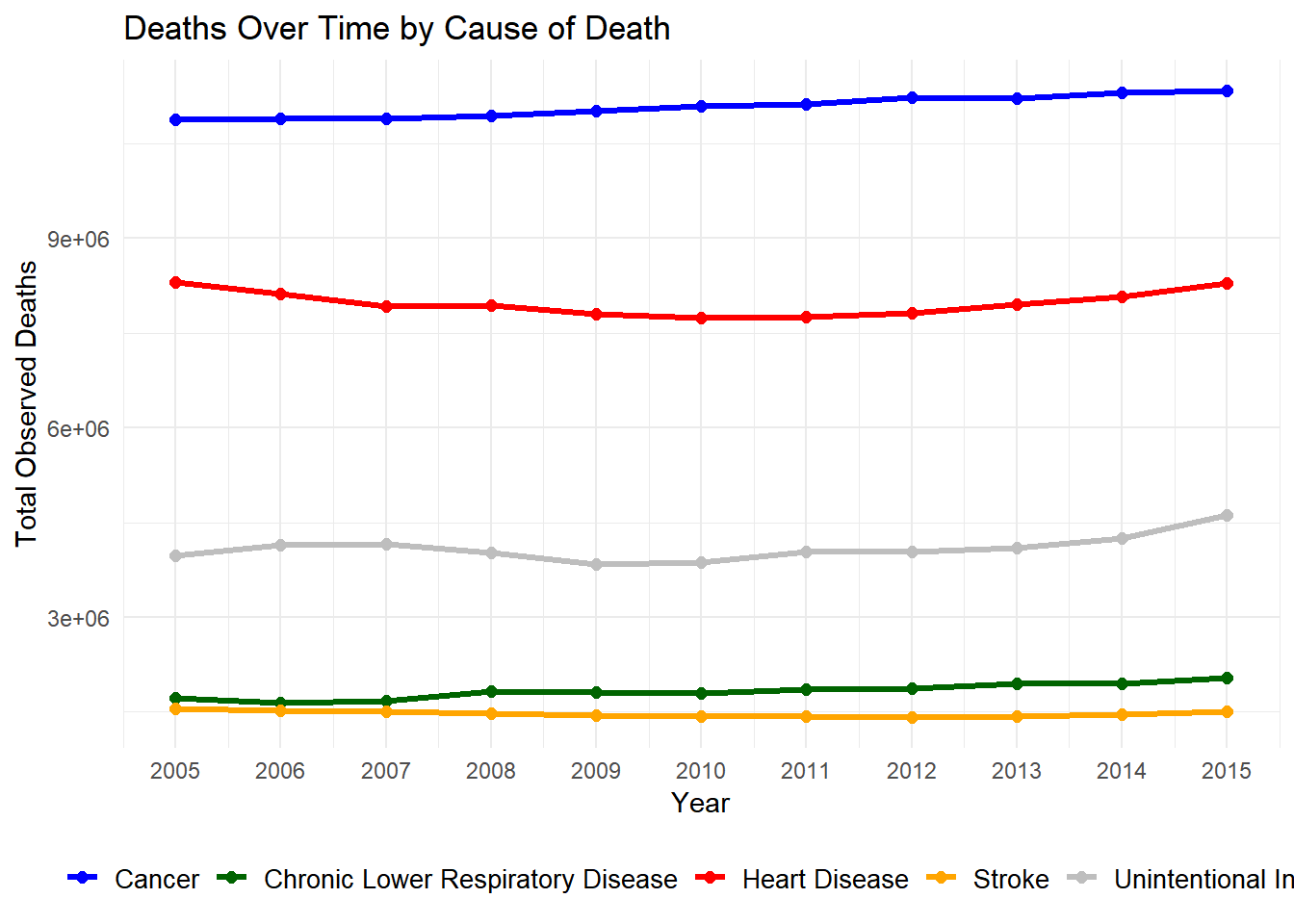
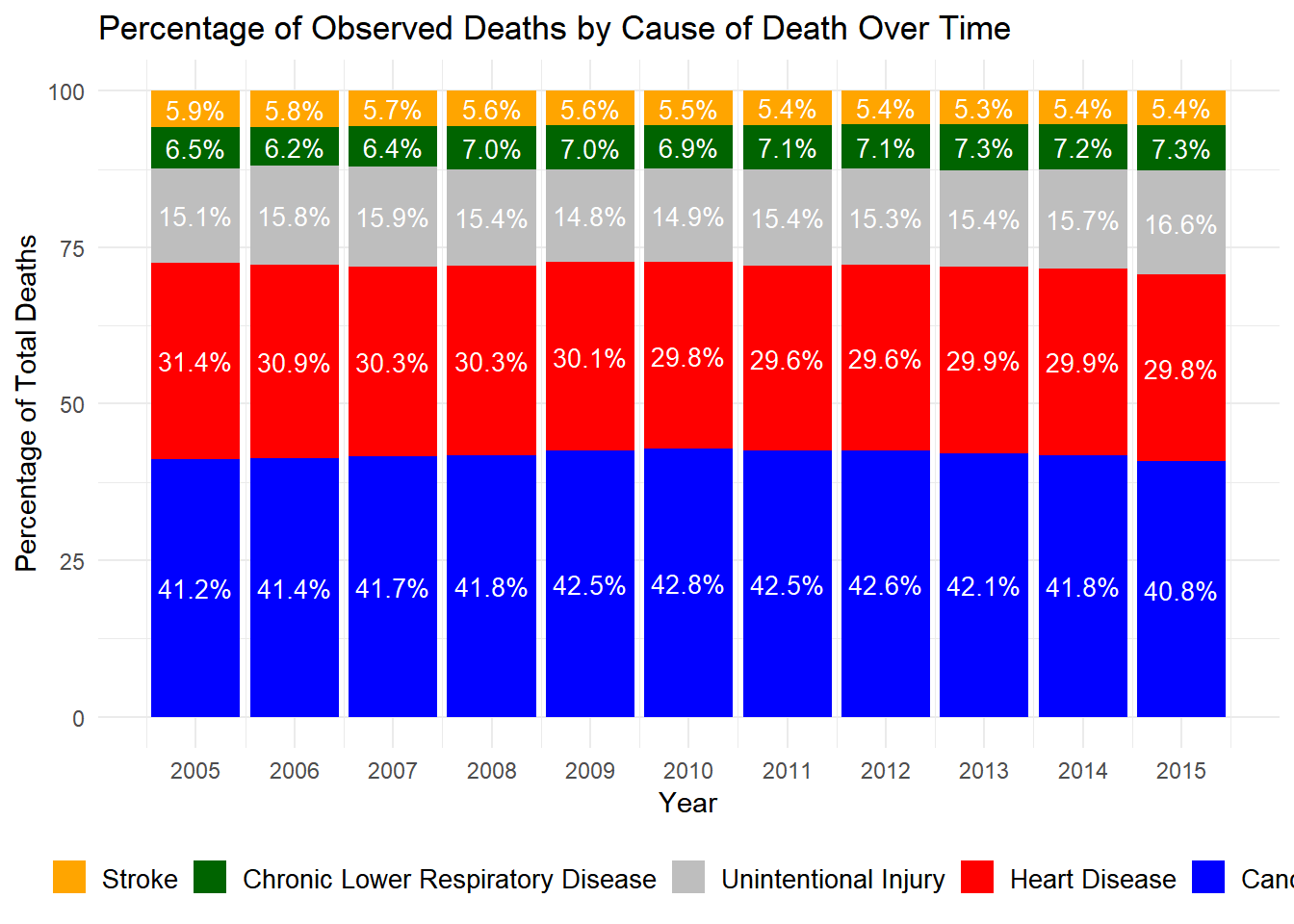
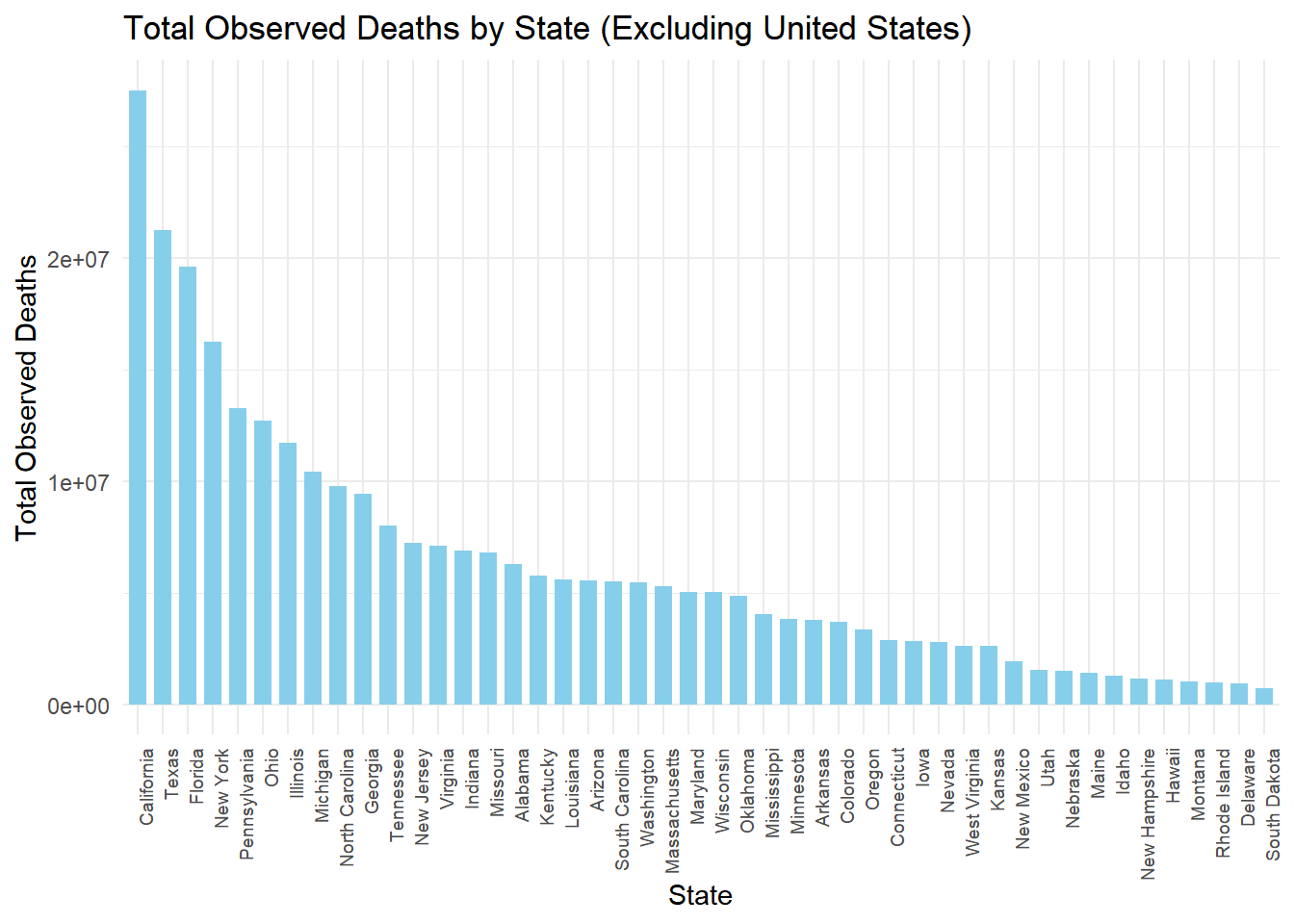
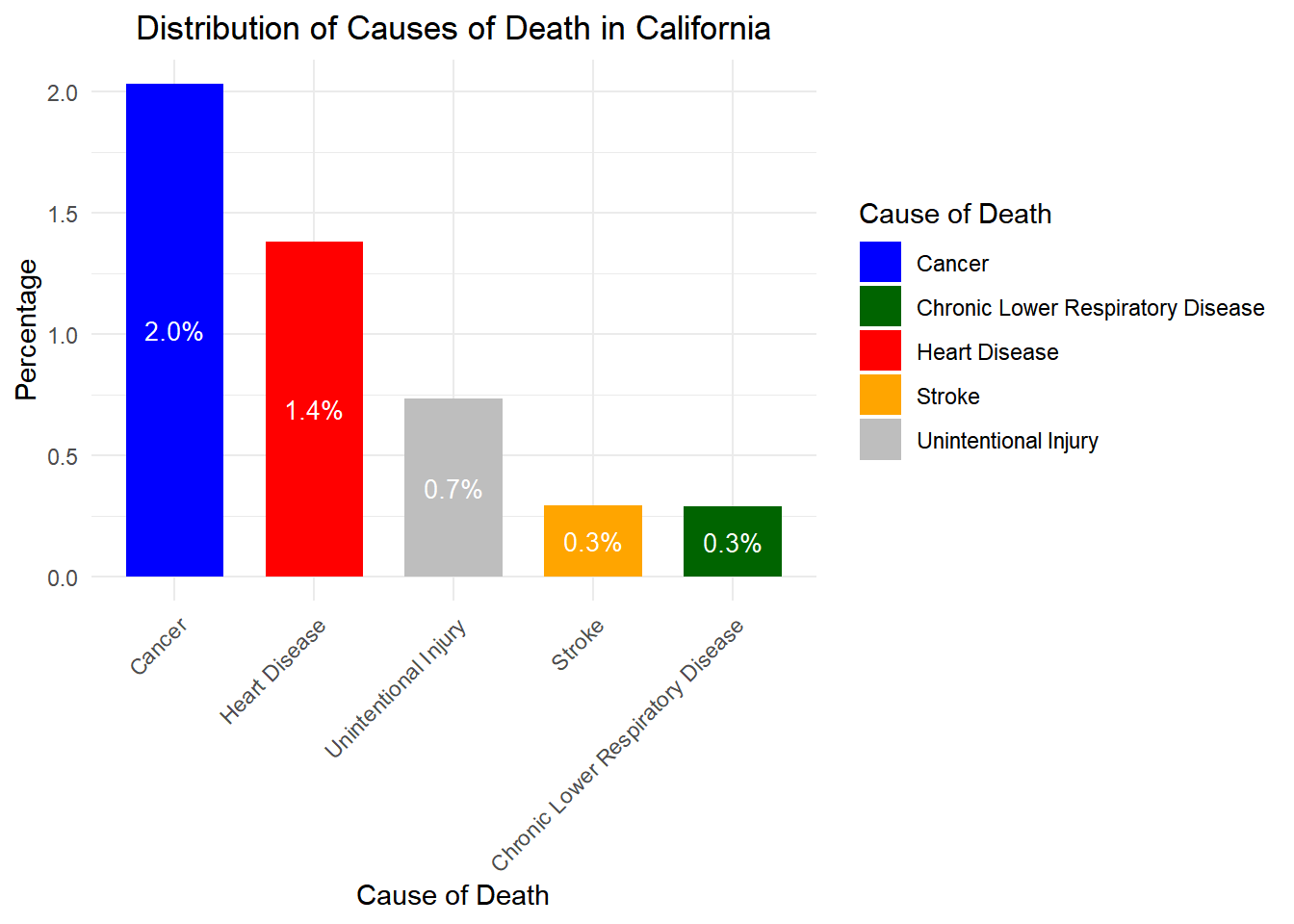
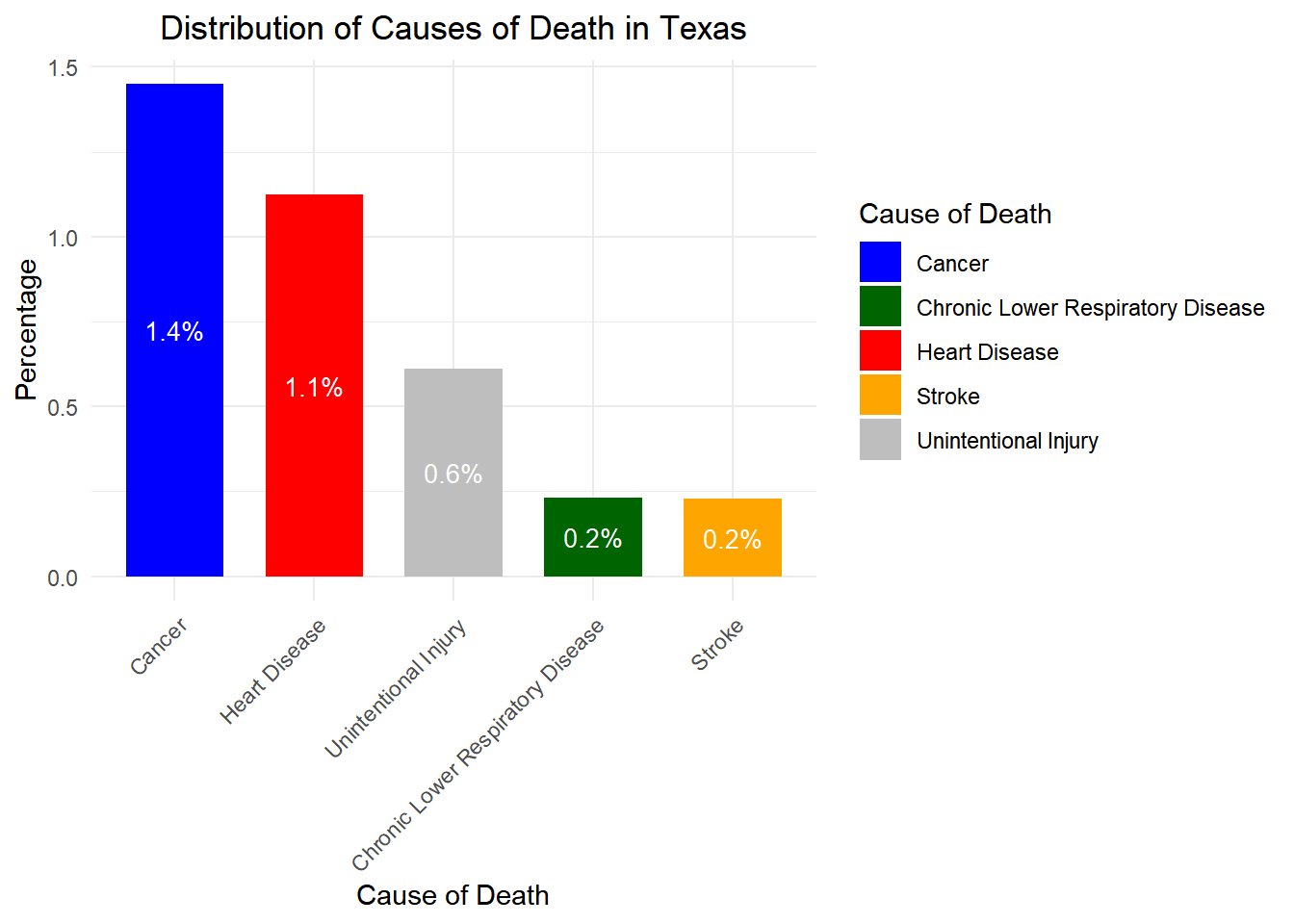
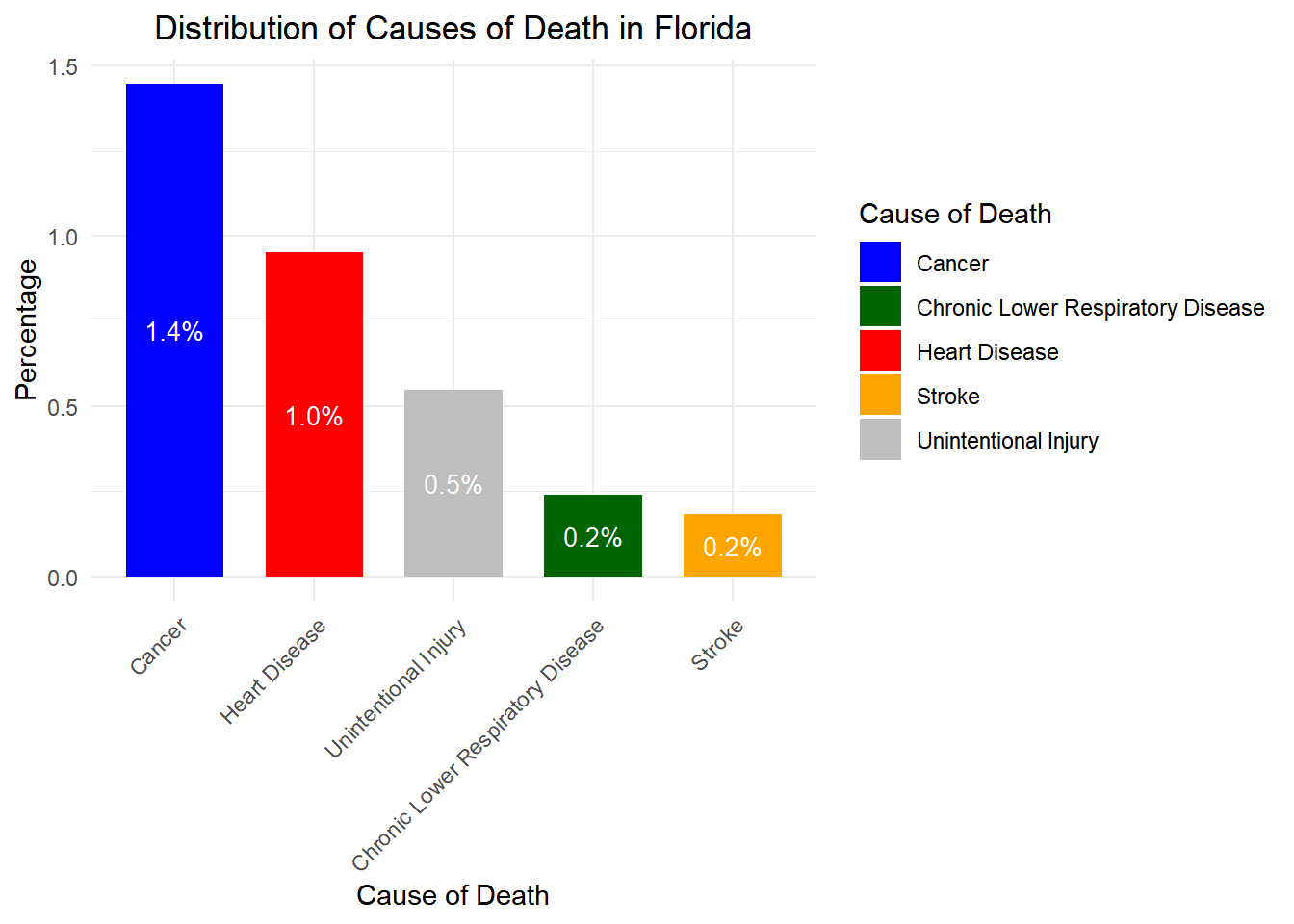
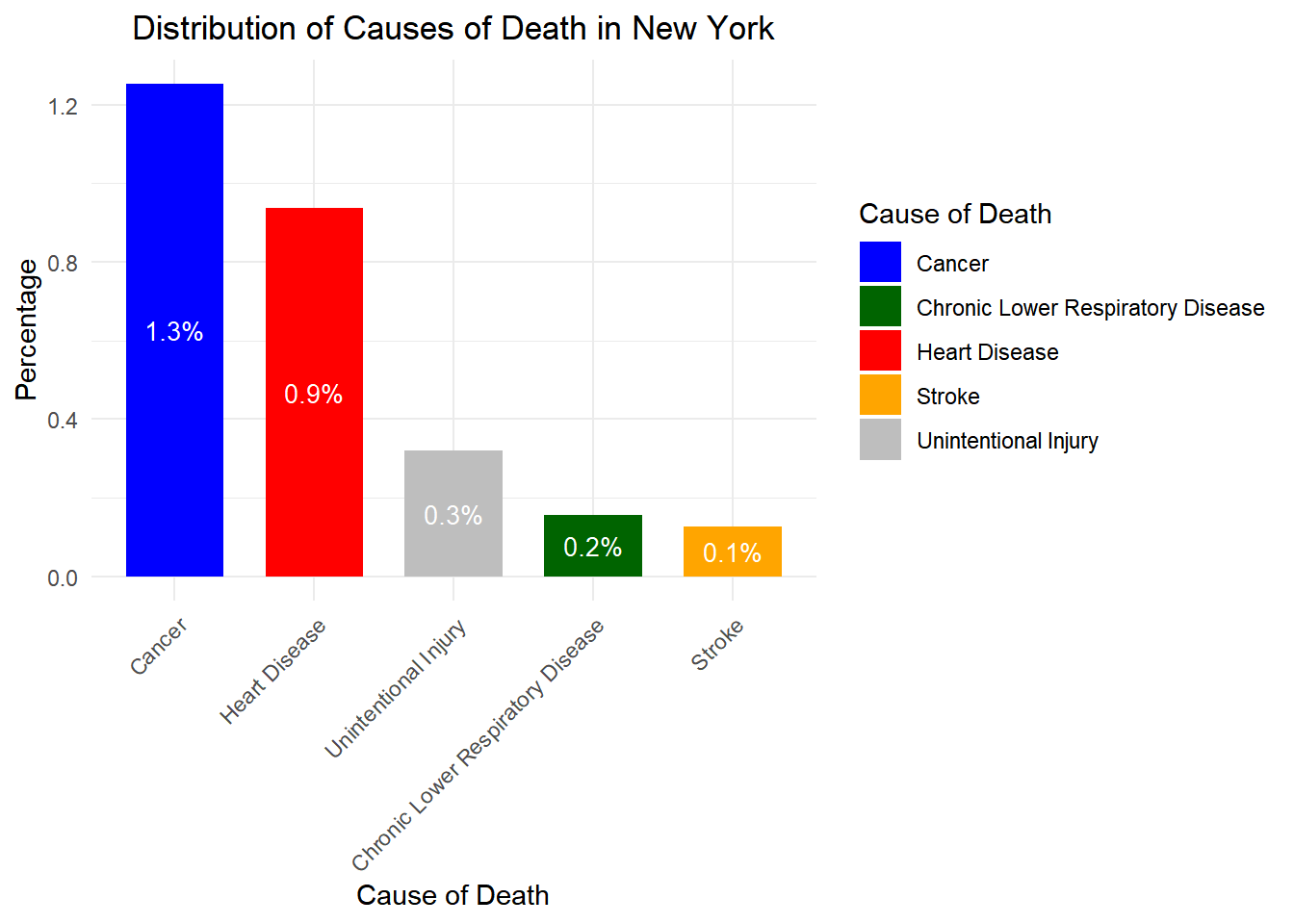
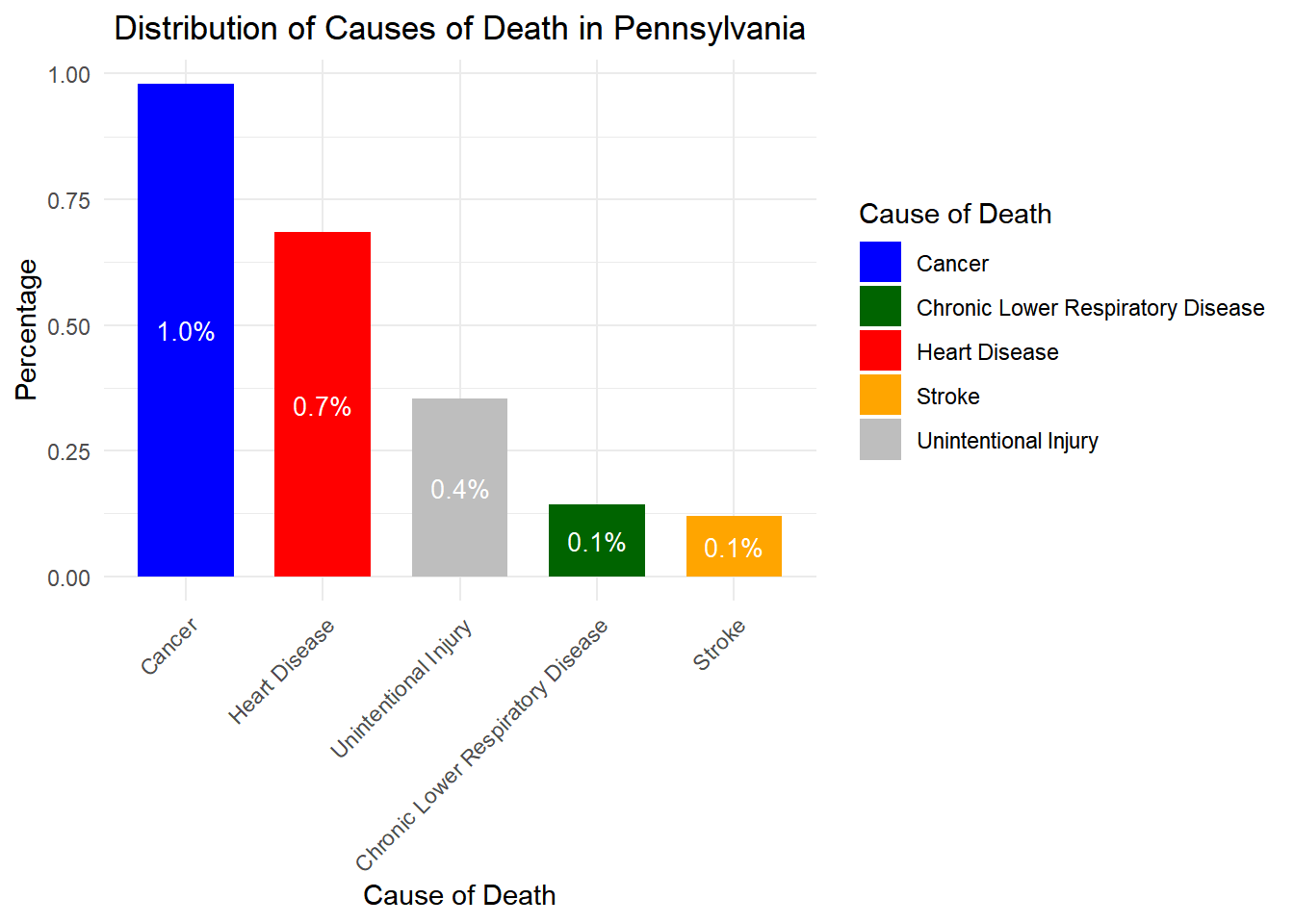
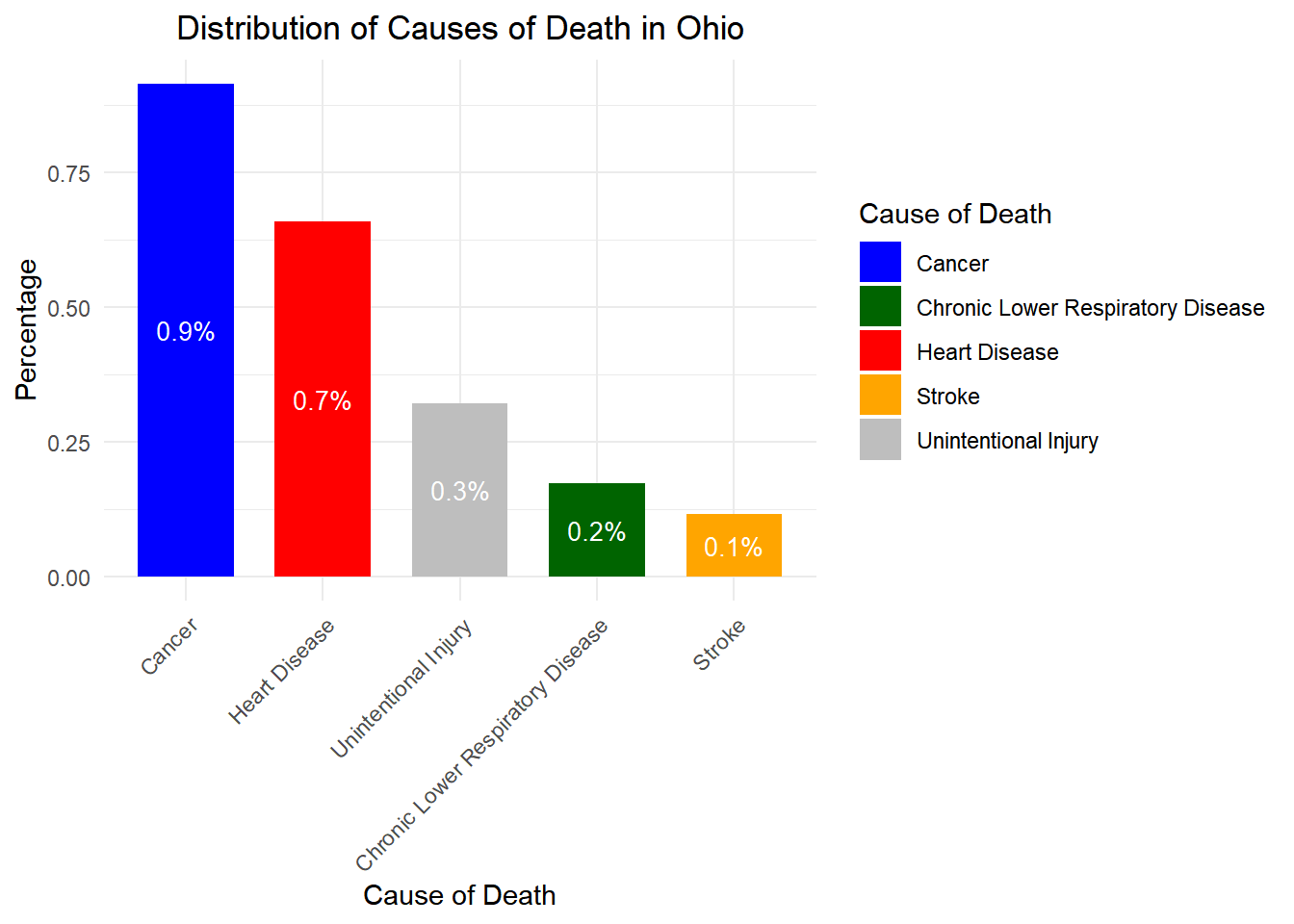
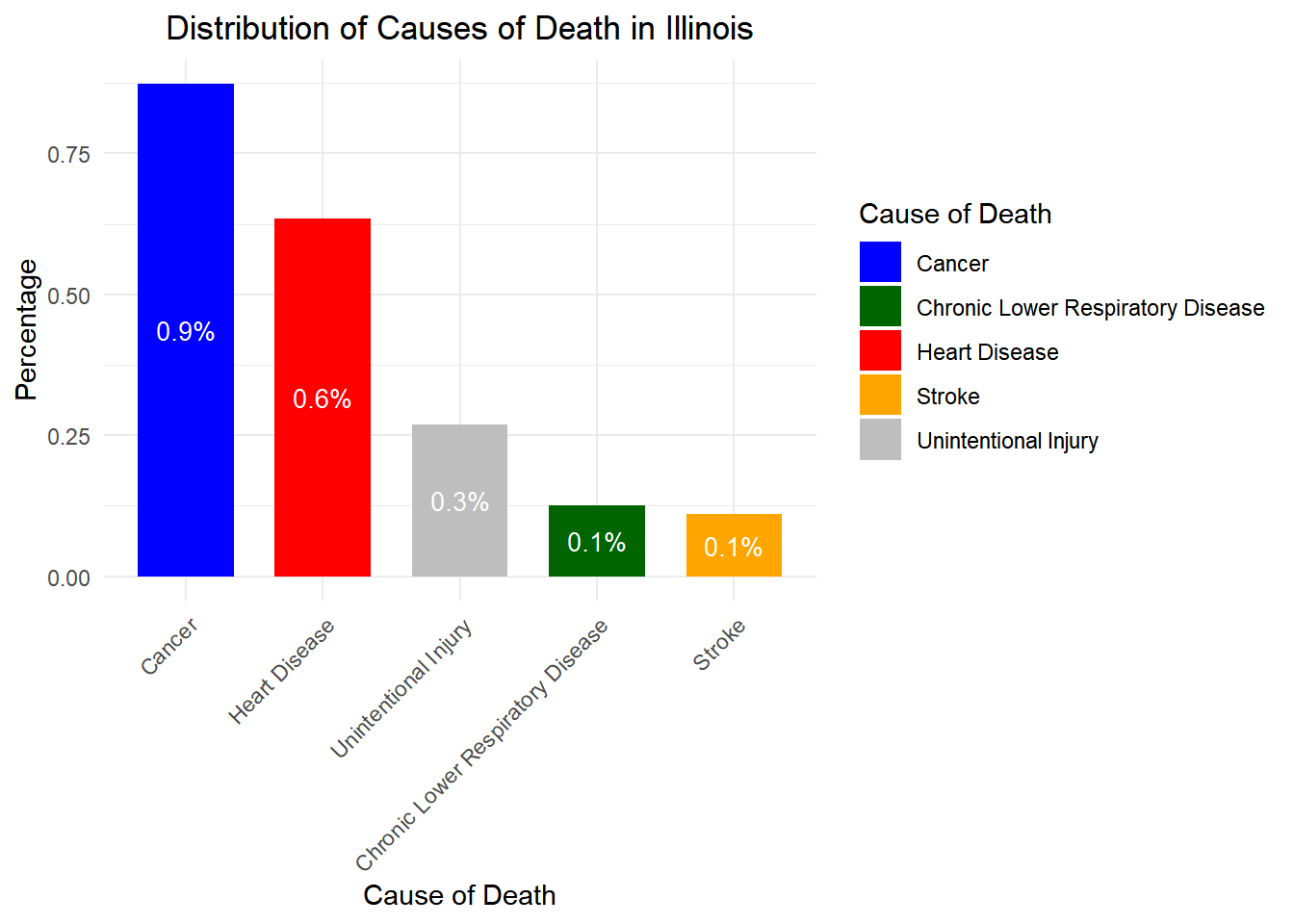
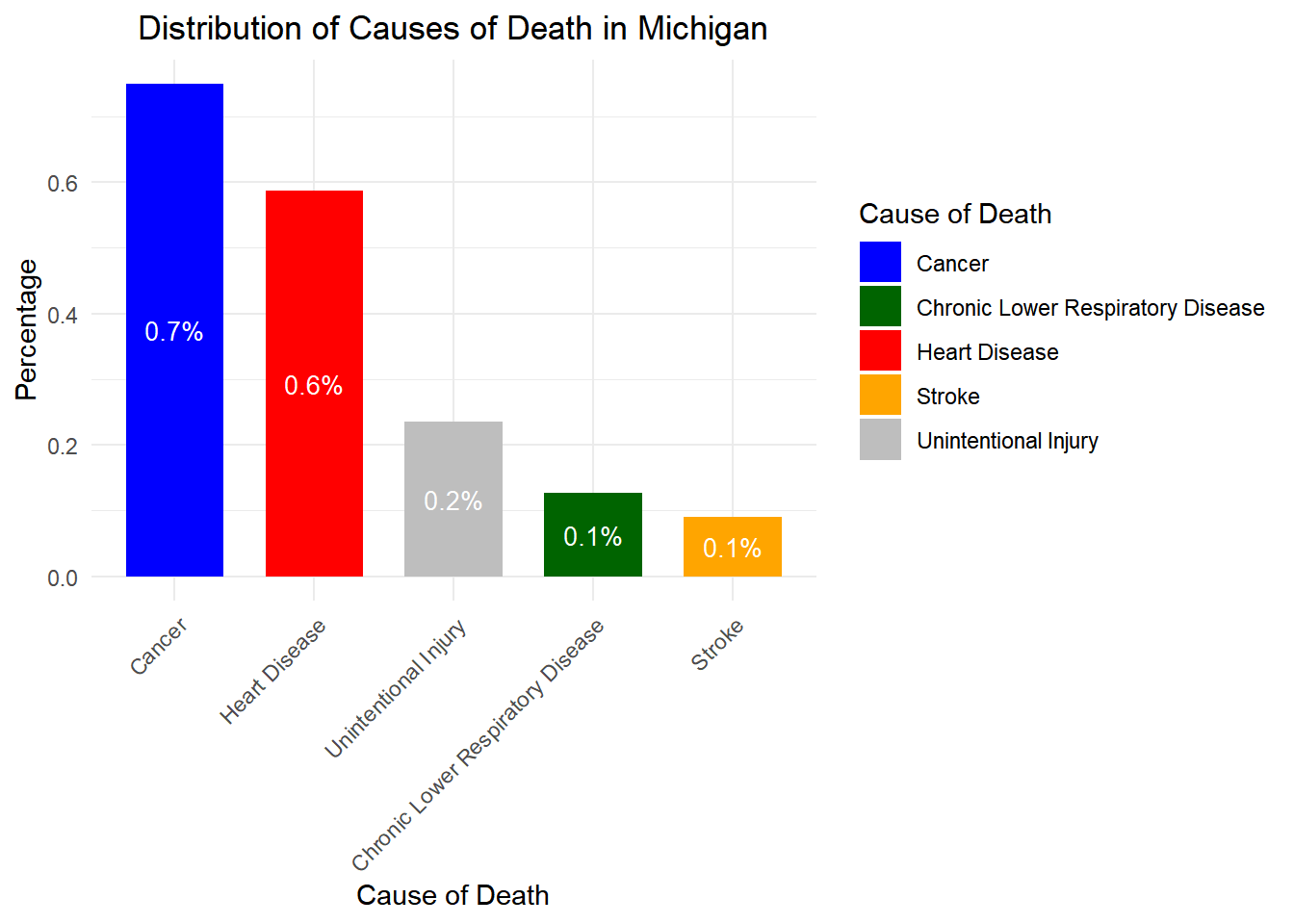
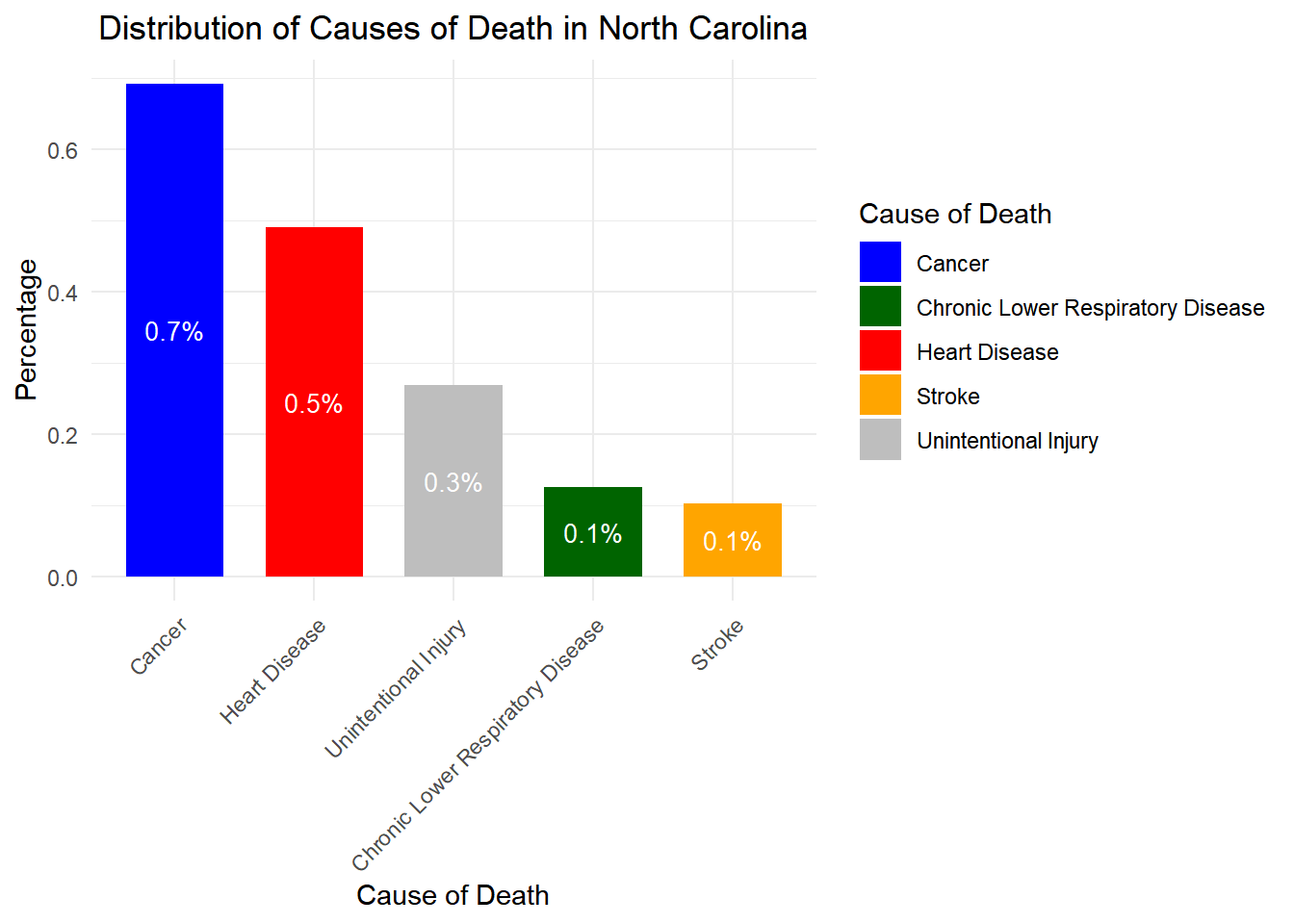
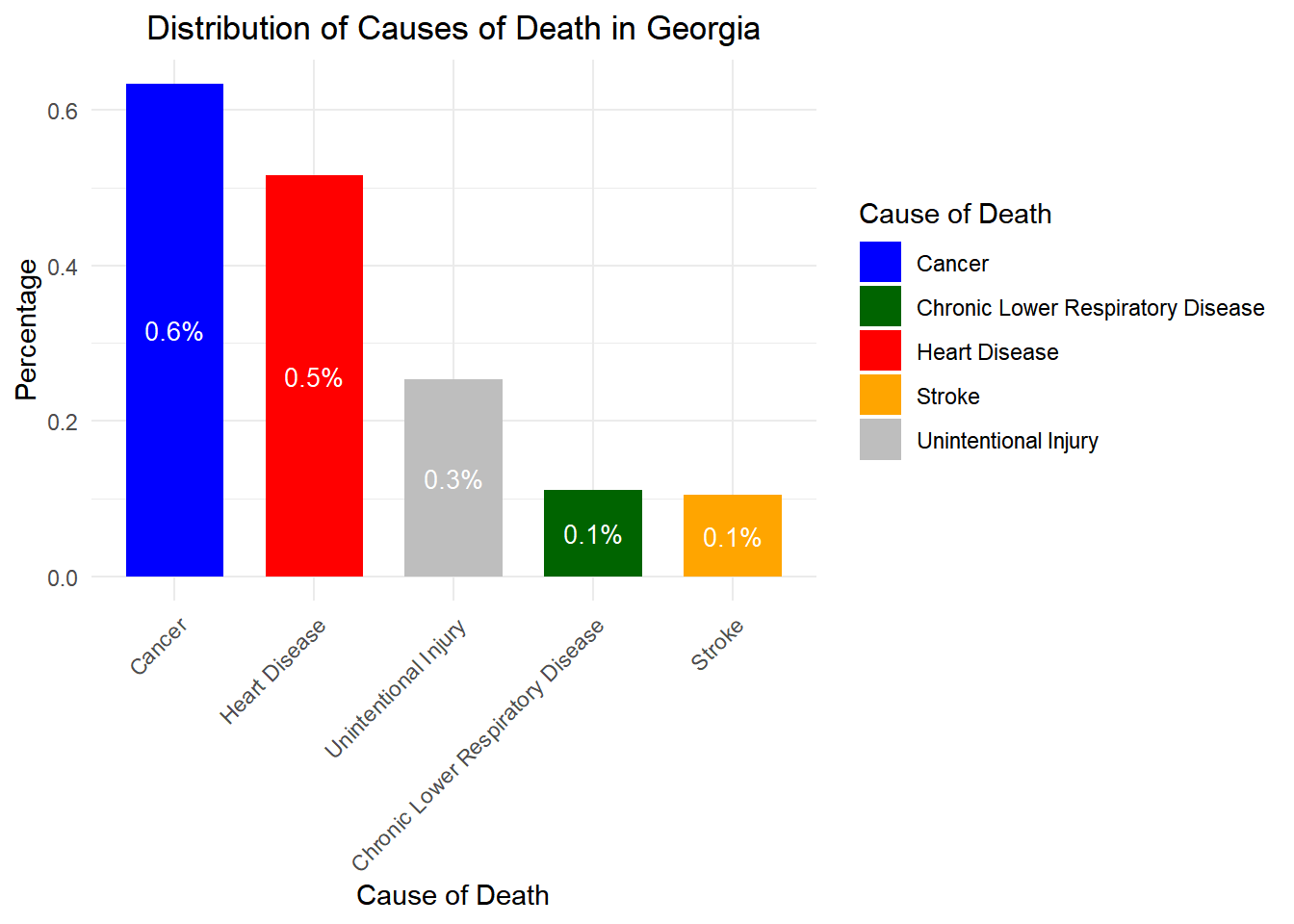
# Load necessary libraries
library(tidyverse)Warning: package 'ggplot2' was built under R version 4.3.3Warning: package 'tidyr' was built under R version 4.3.3Warning: package 'dplyr' was built under R version 4.3.3── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.4
✔ forcats 1.0.0 ✔ stringr 1.5.0
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.2 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorslibrary(readr)
library(RColorBrewer)
library(dplyr)
library(ggplot2)
library(tibble)
# Set the working directory to the project directory
data <- here::here("cdcdata-exercise","NCHS_-_Five_Leading_Causes_of_Death.csv")
# Define the file path and load the data
data <- read_csv("NCHS_-_Five_Leading_Causes_of_Death.csv")Rows: 205920 Columns: 13
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (6): Cause of Death, State, State FIPS Code, Age Range, Benchmark, Locality
dbl (7): Year, HHS Region, Observed Deaths, Population, Expected Deaths, Pot...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.# Tell us the name of the columns
colnames(data) [1] "Year" "Cause of Death"
[3] "State" "State FIPS Code"
[5] "HHS Region" "Age Range"
[7] "Benchmark" "Locality"
[9] "Observed Deaths" "Population"
[11] "Expected Deaths" "Potentially Excess Deaths"
[13] "Percent Potentially Excess Deaths"#output two newline characters
cat("\n\n")# Display the structure of the dataset
str(data)spc_tbl_ [205,920 × 13] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
$ Year : num [1:205920] 2005 2005 2005 2005 2005 ...
$ Cause of Death : chr [1:205920] "Cancer" "Cancer" "Cancer" "Cancer" ...
$ State : chr [1:205920] "Alabama" "Alabama" "Alabama" "Alabama" ...
$ State FIPS Code : chr [1:205920] "AL" "AL" "AL" "AL" ...
$ HHS Region : num [1:205920] 4 4 4 4 4 4 4 4 4 4 ...
$ Age Range : chr [1:205920] "0-49" "0-49" "0-49" "0-49" ...
$ Benchmark : chr [1:205920] "2005 Fixed" "2005 Fixed" "2005 Fixed" "2010 Fixed" ...
$ Locality : chr [1:205920] "All" "Metropolitan" "Nonmetropolitan" "All" ...
$ Observed Deaths : num [1:205920] 756 556 200 756 556 ...
$ Population : num [1:205920] 3148377 2379871 768506 3148377 2379871 ...
$ Expected Deaths : num [1:205920] 451 341 111 421 318 103 451 341 111 784 ...
$ Potentially Excess Deaths : num [1:205920] 305 217 89 335 238 97 305 217 89 562 ...
$ Percent Potentially Excess Deaths: num [1:205920] 40.3 39 44.5 44.3 42.8 48.5 40.3 39 44.5 41.8 ...
- attr(*, "spec")=
.. cols(
.. Year = col_double(),
.. `Cause of Death` = col_character(),
.. State = col_character(),
.. `State FIPS Code` = col_character(),
.. `HHS Region` = col_double(),
.. `Age Range` = col_character(),
.. Benchmark = col_character(),
.. Locality = col_character(),
.. `Observed Deaths` = col_double(),
.. Population = col_double(),
.. `Expected Deaths` = col_double(),
.. `Potentially Excess Deaths` = col_double(),
.. `Percent Potentially Excess Deaths` = col_double()
.. )
- attr(*, "problems")=<externalptr> cat("\n\n")# Check for missing values
colSums(is.na(data)) Year Cause of Death
0 0
State State FIPS Code
0 0
HHS Region Age Range
0 0
Benchmark Locality
0 0
Observed Deaths Population
10212 5280
Expected Deaths Potentially Excess Deaths
10212 10212
Percent Potentially Excess Deaths
10212 cat("\n\n")# Remove rows with NA values
data_cleaned <- data %>%
drop_na()
# Display the structure of the cleaned dataset
str(data_cleaned)tibble [195,708 × 13] (S3: tbl_df/tbl/data.frame)
$ Year : num [1:195708] 2005 2005 2005 2005 2005 ...
$ Cause of Death : chr [1:195708] "Cancer" "Cancer" "Cancer" "Cancer" ...
$ State : chr [1:195708] "Alabama" "Alabama" "Alabama" "Alabama" ...
$ State FIPS Code : chr [1:195708] "AL" "AL" "AL" "AL" ...
$ HHS Region : num [1:195708] 4 4 4 4 4 4 4 4 4 4 ...
$ Age Range : chr [1:195708] "0-49" "0-49" "0-49" "0-49" ...
$ Benchmark : chr [1:195708] "2005 Fixed" "2005 Fixed" "2005 Fixed" "2010 Fixed" ...
$ Locality : chr [1:195708] "All" "Metropolitan" "Nonmetropolitan" "All" ...
$ Observed Deaths : num [1:195708] 756 556 200 756 556 ...
$ Population : num [1:195708] 3148377 2379871 768506 3148377 2379871 ...
$ Expected Deaths : num [1:195708] 451 341 111 421 318 103 451 341 111 784 ...
$ Potentially Excess Deaths : num [1:195708] 305 217 89 335 238 97 305 217 89 562 ...
$ Percent Potentially Excess Deaths: num [1:195708] 40.3 39 44.5 44.3 42.8 48.5 40.3 39 44.5 41.8 ...